
To open and process reaction monitoring data For RM data. Use factor k and define formulas to convert it into reaction times (Z) for subsequent analysis.
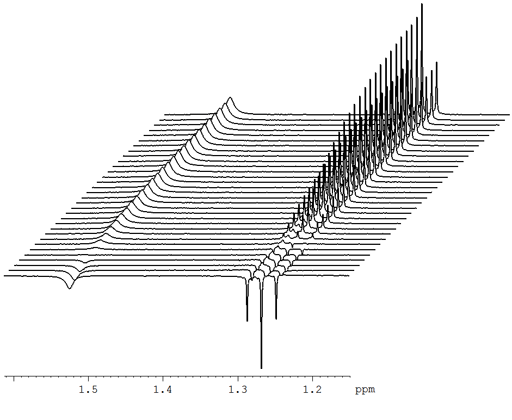
For all these cases, Mnova will only process the acquired dimension (t2). Additionally, Mnova can detect spectra which have been acquired in the so-called arrayed-mode (or pseudo 2D), typically used in relaxation, kinetics or diffusion experiments.One very useful setting for 2D spectra is the appearance of the contou As you did with the 1D spectrum, double-click on the 2D spectrum to customize the appearance. To make Mnova apply linear prediction parameters automatically when it reads data in, go to the settings (upper right corner), go to NMR, then to Import, and be sure that Linear Prediction (under Parameters) is selected.Every object in Mnova can be relocated and resized. Use the green handles to move, rotate and resize the text box. Press Report to report the parameters as a text box on the spectrum. Go to View/Tables/Parameters to view the acquisition parameters.Mestrenova acquisition parameters - mestrenova quick guid For more info on IUPAC recommendations the facility web site. Absolute referencing in MestreNova allows to reference X-nucleus and 2D-spectra based on a correctly referenced 1H spectrum using the absolute frequency of the TMS signal.2D spectra processing FT Bruker : xfb Varian : wft2da. MestReNova Version 12.0.3 Acquisition Parameter NS : Number of Scan SW : Spectral Width O1P, O2P : Offset TD : Number of point NUS Experiment Homonuclear : COSY, NOESY, ROESY, TOCSY Heteronuclear : HSQC, HMBC, H2BC, Non-uniform Sampling Save Time for 2D acquisition 25 ~ 50% Uniform sampling Non -Uniform Sampling NUS processing.Resize the text box and spectrum to make a better layout Use the green handles to move, rotate and resize the text bo To see the parameters Choose View | Tables | Parameters to view the acquisition and processing parameters Click Report to report the parameters as a text box on the spectrum.The user will be able to obtain any parameter from any raw data file (procs, acqus, title, etc.) by selecting the desired file on the 'File/Block' box (red square in the.

Clicking on the 'More>' button will display the 'Current Spectrum Parameters' window, as you can see in the picture below. Customization of the parameters table is Mnova is very intuitive, just follow the menu: View/Tables/Parameters and click on the 'Customize' icon to display the 'Customize Parameters' dialog box.The userRead Mor FYI: you can display the acquisition and processing parameters if you like. The user can select the properties of the spectrum by following the menu 'Edit/Properties' or by double clicking the left mouse button or pressing the right mouse button on the spectrum display and selecting 'Properties' from the pop-up menu. Mnova will allow the user to customize nearly all the attributes of the spectrum. Auf die Plätze, fertig & loslernen! Anschauliche Lernvideos, vielfältige Übungen und hilfreiche Arbeitsblätter Mestrenova acquisition parameters Spectrum Properties - Mestrelab Resource.


 0 kommentar(er)
0 kommentar(er)
